
Host Onboarding
Unlock the potential of non-model microbes
Non-model microbes have unique phenotypes that can be beneficial for biomanufacturing, but typically lack foundational datasets and basic genetic tools needed for rational strain development. Our host onboarding capabilities include the rapid development of tools and datasets that allow us to unlock the potential of non-model microbes.
Host development evaluation
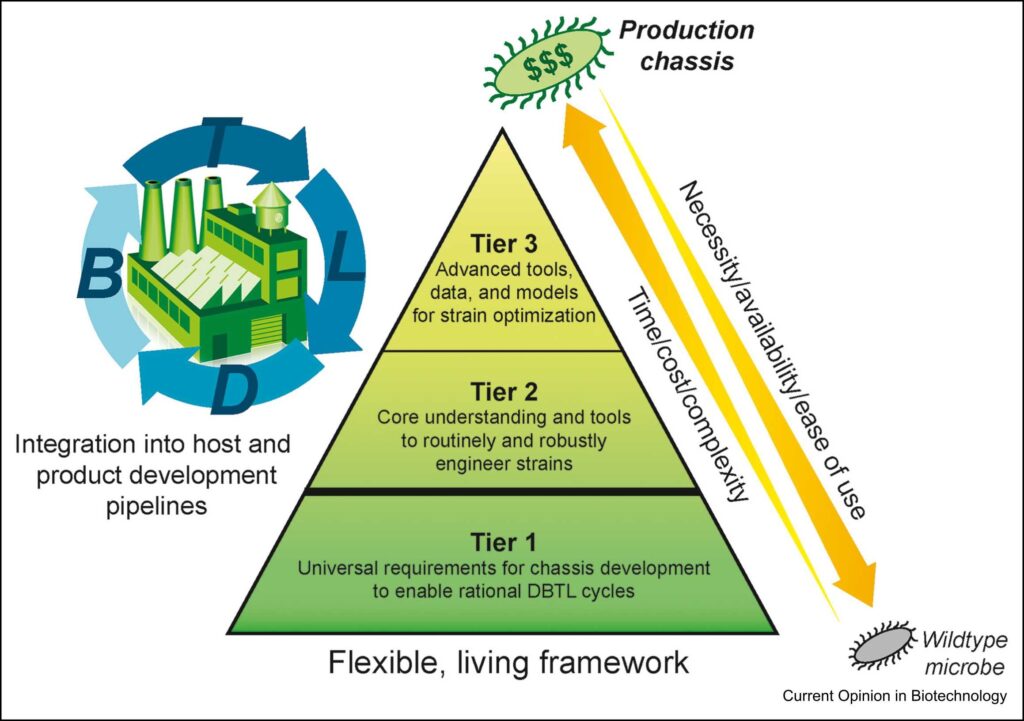
Our Tier System organizes, tracks, and prioritizes our strain development efforts. Our initial focus is on developing genetic tools for creating chassis strains for metabolic engineering. Advanced tiers include omics datasets, fermentation data, advanced genetic tools, and models to support host development. Progress on Agile BioFoundry host organisms is tracked through the Host Onboarding Tool (HObT) website.
Genetic tool development
Rapid development of genetic tools is essential for rational strain engineering. We have a growing collection of genetic parts for diverse bacteria and fungi, which are adaptable to new organisms. Our transformation methods, chromosomal modification methods, gene expression “parts”, and high throughput tools and protocols accelerate bioengineering through the Design-Build-Test-Learn process.
Datasets
Large datasets provide key insights needed for Learn efforts. We can perform transcriptomics, proteomics, and metabolomics to help understand gene expression and metabolic flux. We can also characterize growth and bioconversion across scales from microtiters to thousands of liters. Our genetic tools enable the construction of genome-scale libraries of engineered strains and large libraries of pathway variants that can help us understand and improve strain performance. These datasets will be made publicly available through HObT to accelerate the development and adoption of these strains by academia and industry.
Currently, onboarded microbes within the Agile BioFoundry include:
- Aspergillus niger
- Aspergillus pseudoterreus
- Bacillus licheniformis
- Clostridium ljungdahlii
- Clostridium tyrobutyricum
- Corynebacterium glutamicum
- Cupriavidus necator
- Lipomyces starkeyi
- Pichia kudriazevii
- Pseudomonas fluorescens
- Pseudomonas putida
- Rhodobacter sphaeroides
- Rhodosporidium toruloides
- Yarrowia lipolytica
- Zymomonas mobilis