
Machine Learning
Predicting bioengineered systems to drive metabolic engineering
We often don’t know all the biological mechanisms required to predict the behavior of a bioengineered system through mechanistic modeling.
In those cases, a data-intensive, statistics-based approach can predict bioengineered systems to the degree required to drive metabolic engineering efforts.
We use a variety of machine learning approaches that range from deep learning to our own methods developed specifically for synthetic biology. We have successfully used these techniques to guide metabolic engineering in an effective fashion.
Our machine learning techniques are coupled with our ability to produce our own data in an automated fashion using our capabilities.
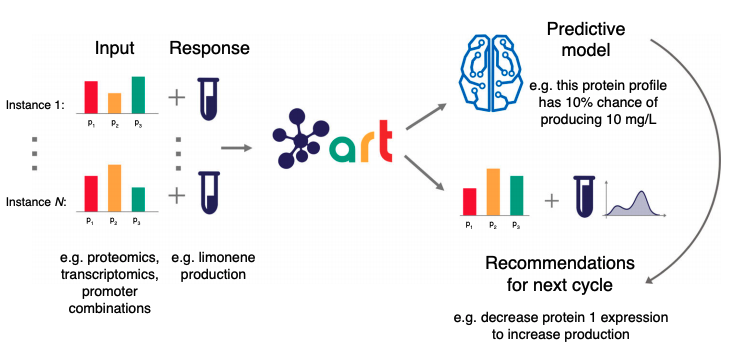
Machine learning provides a systematic method to leverage data stored in the Experiment Data Depot (EDD) to guide metabolic engineering methods without the need for a deep mechanistic understanding.
This data-intensive approach is usually limited by the availability of data. Typically, 50-100 conditions (strains) are needed for successful predictions, as well as 3-4 consecutive rounds in which predictions are tested and the new data is used to improve predictions for the next round.
Labs:
- Argonne National Laboratory
- Lawrence Berkeley National Laboratory
- Pacific Northwest National Laboratory
- National Renewable Energy Laboratory