
Computational Protein Design
Our suite of protein modeling tools perform structure based design of enzymes for gain-of-function.
Natural enzymes are efficient, but can lack an optimal catalytic efficiency, stability and undergo product inhibition when applied in an industrial set up. These enzymes may become bottlenecks in metabolic pathway engineering efforts. Increasing the expression of the enzyme by using a strong promoter or achieving gain-of-function using error prone PCR and adaptive laboratory evolution can help alleviate the bottleneck.
Alternatively, the rational design approach using ROSETTA can predict mutations that can enhance stability, reduce product inhibition or improve catalytic efficiency of enzymes. This approach takes advantage of a large structural database to predict structure from the sequences and identify a very focused region on the protein to mutate in order to improve stability or catalytic efficiency of the enzyme.
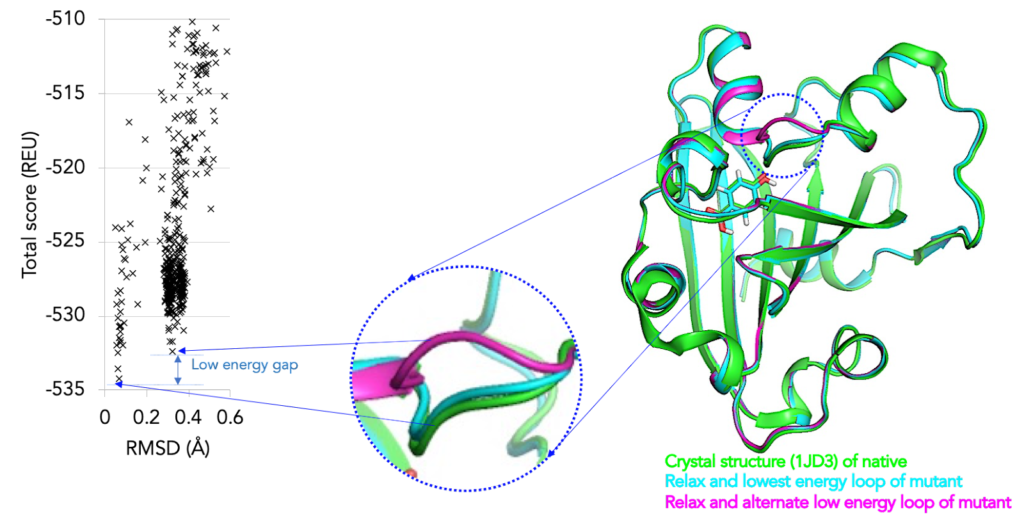
While computational protein modeling tools help eliminate the majority of the substitutions that are most likely destructive to the protein, it also provides multiple solutions to a given scientific problem and can help inform a small set of promising mutations which can be combinatorially built and tested using other capabilities such as “Smart” Microbial Cell Technology or Microfluidic Biocatalyst optimization.