
Artificial Intelligence
Guide metabolic engineering efforts, make predictions, and optimize production
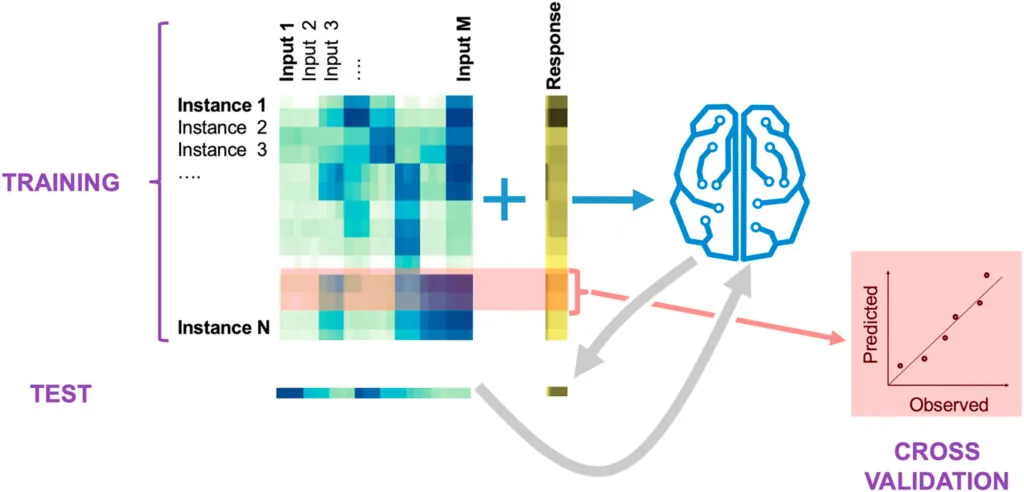
We use a variety of machine learning (ML) approaches developed specifically for synthetic biology.
These approaches, combined with our ability to produce data in an automated and high-throughput fashion, allow us to guide metabolic engineering in an effective manner through active learning processes — a type of machine learning where an algorithm decides which experiments to perform next.
Using this type of machine learning, we can systematically leverage data stored in a standard ontology data repository (Experiment Data Depot) and guide metabolic engineering efforts without a deep mechanistic understanding of the biological systems involved.
ART
The Automated Recommendation Tool (ART) is a general-purpose machine learning tool that has been successfully used to inform and guide synthetic biology. ART has been used to predict the production of the renewable biofuel precursor limonene from proteomics data, predict the proteins that would result in a particular beer taste profile, recommend promoters that improve tryptophan productivity, design media to increase flaviolin production, and predict which genes to downregulate through CRISPRi for improved titers.
ART is tailored to the synthetic biology needs: small data sets (obtained results with as little as 27 instances), the need for uncertainty quantification, and recursive Design-Build-Test-Learn cycles (providing recommendations, not just predictions).
Kinetic Learning
Our kinetic learning method combines machine learning and multi-omics data to effectively predict pathway dynamics in an automated fashion. This method outperforms kinetic models and makes predictions that can productively guide bioengineering efforts.
Regulatory Deep Learning
Our deep learning methods connect microbes’ natural information processing and environmental sensing abilities to the predictive computational design of regulatory networks. As a result, microbes become fully programmable biosystems capable of bioprocesses that are difficult to achieve otherwise.
USE CASE
We used promoter and production data to train machine learning algorithms that recommended new strain designs that improved tryptophan productivity by 106% over the base strain.
Our approach used a variety of tools to effectively leverage the growing amount of synthetic biology data now available.
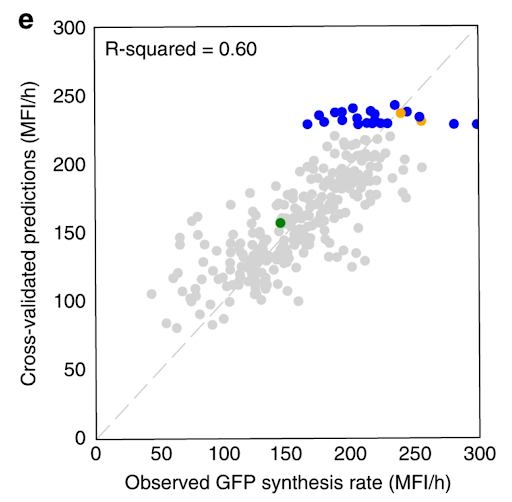
Our Automated Recommendation Tool (ART) provided predictions (in gray) that closely match actual measurements of tryptophan productivity, as shown in cross-validated predictions. Recommendations of new promoter combinations (in blue) eventually improved tryptophan productivity by 106% than the base strain (in green).
Publications
- A machine learning Automated Recommendation Tool for synthetic biology
- A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data
- Combining mechanistic and machine learning models for predictive engineering and optimization of tryptophan metabolism
- Machine learning-led semi-automated medium optimization reveals salt as key for flaviolin production in Pseudomonas putida
- Machine learning for metabolic engineering: A review
- Multiomics Data Collection, Visualization, and Utilization for Guiding Metabolic Engineering