
Build your bioprocess.
We partner with the biomanufacturing industry to accelerate bioproduct development.
New Strategic Core Industry Partnership Opportunity
Leverage the Agile BioFoundry’s cutting-edge facilities, capabilities, and expertise.

Advancing biomanufacturing across the public and private sectors
The Agile BioFoundry is a consortium of national laboratories dedicated to accelerating biomanufacturing and decarbonizing the economy.
We collaborate with industry and academia to rapidly innovate the development of bio-based products.
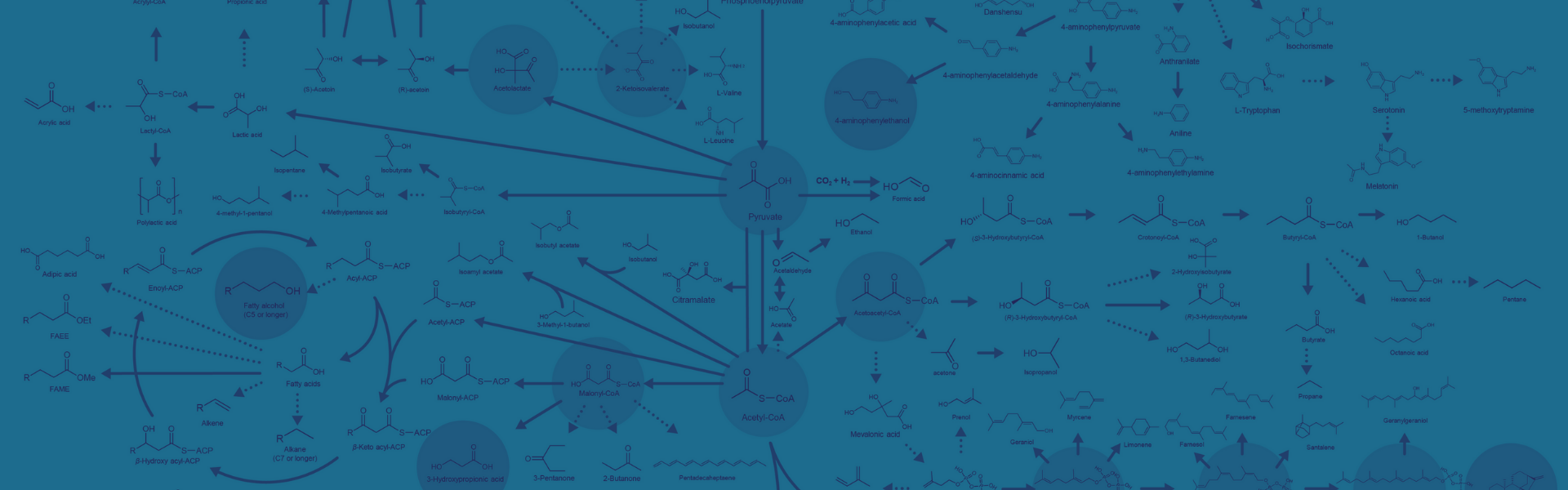
Get where you want to go, faster.
Discover how the Agile BioFoundry can get you closer to your target.
We unite the unique abilities of the national laboratories through a Design-Build-Test-Learn platform.
News and Events
Agile BioFoundry Partners with Pow.Bio to Test Continuous Fermentation Platform
March 27, 2024
The Agile BioFoundry is partnering with Pow.Bio to test their continuous fermentation platform in a promising microbial host.
Agile BioFoundry Announces A New Industry Collaboration Opportunity
February 27, 2024
Leverage the Agile BioFoundry’s cutting-edge facilities, capabilities, and expertise.
Agile BioFoundry Hosts 2nd U.S.-Korea Synthetic Biology Conference
December 07, 2023
The goal of the conference was to facilitate international collaboration between the U.S. and Korea and discuss global synthetic biology standardization and norms.

Help us innovate in synthetic biology and biomanufacturing.
Our state-of-the-art capabilities are available to researchers in the public and private sectors.
Have an idea? Learn more about how to work with us, or get in touch with our team today.






































